Viruses have lots of sneaky tricks they employ to increase the chance of infecting their hosts. Scientists at the University of British Columbia (UBC) have identified a virus that has a particularly sophisticated way of infecting its host, using DNA stolen from a previous host, worthy of being a plot device in a spy movie.
Cyanophage N1 is a type of bacteriophage virus that infects important families of freshwater bacteria, a.k.a cyanobacteria, called Nostoc and Anabaena. It improves its chances of achieving a successful infection, enabling it to reproduce, by utilising a CRISPR DNA sequence that it must have originally stolen from one of its hosts.
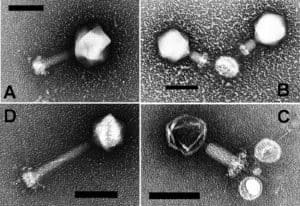
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, which are DNA sequences that evolved in bacteria to provide protection against infection by viruses or plasmids. They do this by recognising DNA sequences within the invader which are then cleaved in two with specific enzymes, thus deactivaing them. So CRISPR sequences basically act like an immune system for bacteria.
However since 2012 CRISPR sequences have also been used as a genome engineering and editing tool, enabling precise and reliable cutting and splicing of DNA in cultured cells or laboratory organisms.
The American Association for the Advancement of Science voted the use of CRISPR in gene editing as the breakthrough of the year in 2015, due to it providing a major step change in the accuracy, ease and cost of gene editing.
Curtis Suttle, lead scientist of the cyanophage study published in mBio, said:
This is the first evidence we’ve seen that a virus can donate an immunity system via CRISPR… This is like a hacker compromising a computer system, and then immediately patching it to ensure other hackers can’t break in.
Suttle and his co-authors think that the cyanophage N1 virus uses the stolen CRISPR sequence to help Nostoc and Anabaena cyanobacteria to fight off infection by other viruses. While still allowing the infected bacteria to be a stable host for the N1 virus, thus increasing its likelihood of successfully reproducing.
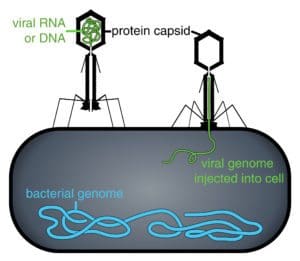
The Nostoc and Anabaena cyanobacteria hosts of the N1 virus are important members of habitats that include: rice paddy soils, ice covered polar lakes, hypertrophic coastal lagoons and rock pool communities in Karst regions.
Ancestors of Nostoc cyanobacteria (cyanobacteria are capable of photosynthesis) were responsible for providing oxygen to the early Earth’s (around 2.3 billion years ago) atmosphere, in what is known as the Great Oxygenation Event (GOE). For anyone who doubts our current climate change is man made, the GOE is an excellent example of how living organisms can change the atmosphere and the climate in a cataclysmic manner.
The cyanobacteria also have a long history with their viruses, according to Suttle:
Bacteria and their viruses have a shared evolutionary history stretching for billions of years… So at some point along the way N1 stole a defensive CRISPR array from Nostoc or a close relative.
Today close relatives of Nostoc are still producing a huge amount of the oxygen we breath. So understanding how cyanobacteria interact with viruses and the environment is important in understanding the maintenance of oxygen levels within our atmosphere, and possibly avoiding another cataclysmic change to our atmosphere.
Featured Image via Wikimedia Commons – Phi X 174 baceriophage which was the first DNA based genome to be sequenced by Fred Sanger and his team in 1977